This vignette gives a demonstration of the package on emulating the popular motorcycle dataset (Silverman 1985).
Load packages and data
We start by loading packages:
We now load the training data points,
X <- mcycle$times
Y <- mcycle$accelscale them,
and plot them:
ggplot(data = data.frame(X = X, Y = Y), aes(x = X, y = Y)) +
geom_point(shape = 16, size = 3) +
labs(x = "Time", y = "Acceleration") +
theme(axis.title = element_text(size = 13),
axis.text = element_text(size = 13))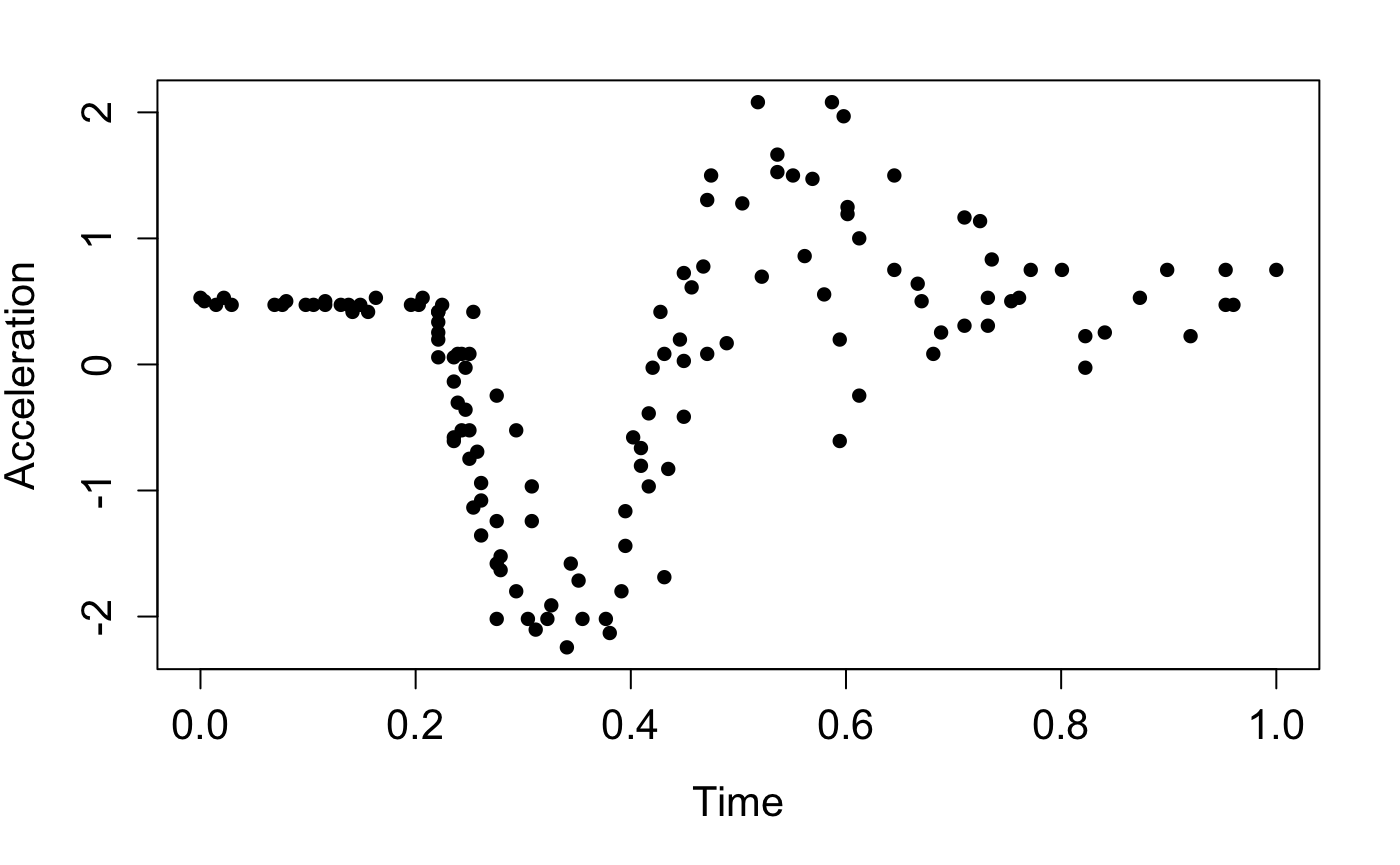
Before constructing an emulator, we first specify a seed with
set_seed() from the package for reproducibility
set_seed(9999)and split a training data set and a testing data set:
Construct and train a DGP emulator
We consider a three-layered DGP emulator with squared exponential kernels and heteroskedastic likelihood:
m_dgp <- dgp(train_X, train_Y, depth = 3, likelihood = "Hetero")## Auto-generating a 3-layered DGP structure ... done
## Initializing the DGP emulator ... done
## Training the DGP emulator:
## Iteration 500: Layer 3: 100%|██████████| 500/500 [00:09<00:00, 53.85it/s]
## Imputing ... doneWe choose a heteroskedastic Gaussian likelihood by setting
likelihood = "Hetero" since the data drawn in the plot show
varying noise. We can use summary() to visualize the
structure and specifications for the trained DGP emulator:
summary(m_dgp)For comparison, we also build a GP emulator (by gp())
that incorporates homogeneous noise by setting
nugget_est = T and the initial nugget value to
:
m_gp <- gp(train_X, train_Y, nugget_est = T, nugget = 1e-2) ## Auto-generating a GP structure ... done
## Initializing the GP emulator ... done
## Training the GP emulator ... doneWe can also summarize the trained GP emulator by
summary():
summary(m_gp)Validation
We are now ready to validate both emulators via
validate() at the 20 out-of-sample testing positions
generated earlier:
m_dgp <- validate(m_dgp, test_x, test_y)## Initializing the OOS ... done
## Calculating the OOS ... done
## Saving results to the slot 'oos' in the dgp object ... done
m_gp <- validate(m_gp, test_x, test_y)## Initializing the OOS ... done
## Calculating the OOS ... done
## Saving results to the slot 'oos' in the gp object ... doneNote that using validate() before plotting can save
subsequent computations compared to simply invoking plot(),
as validate() stores validation results in the emulator
objects and plot() will use these, if it can, to avoid
calculating them on the fly. Finally, we plot the OOS validation for the
GP emulator:
plot(m_gp, test_x, test_y)## Validating and computing ... done
## Post-processing OOS results ... done
## Plotting ... done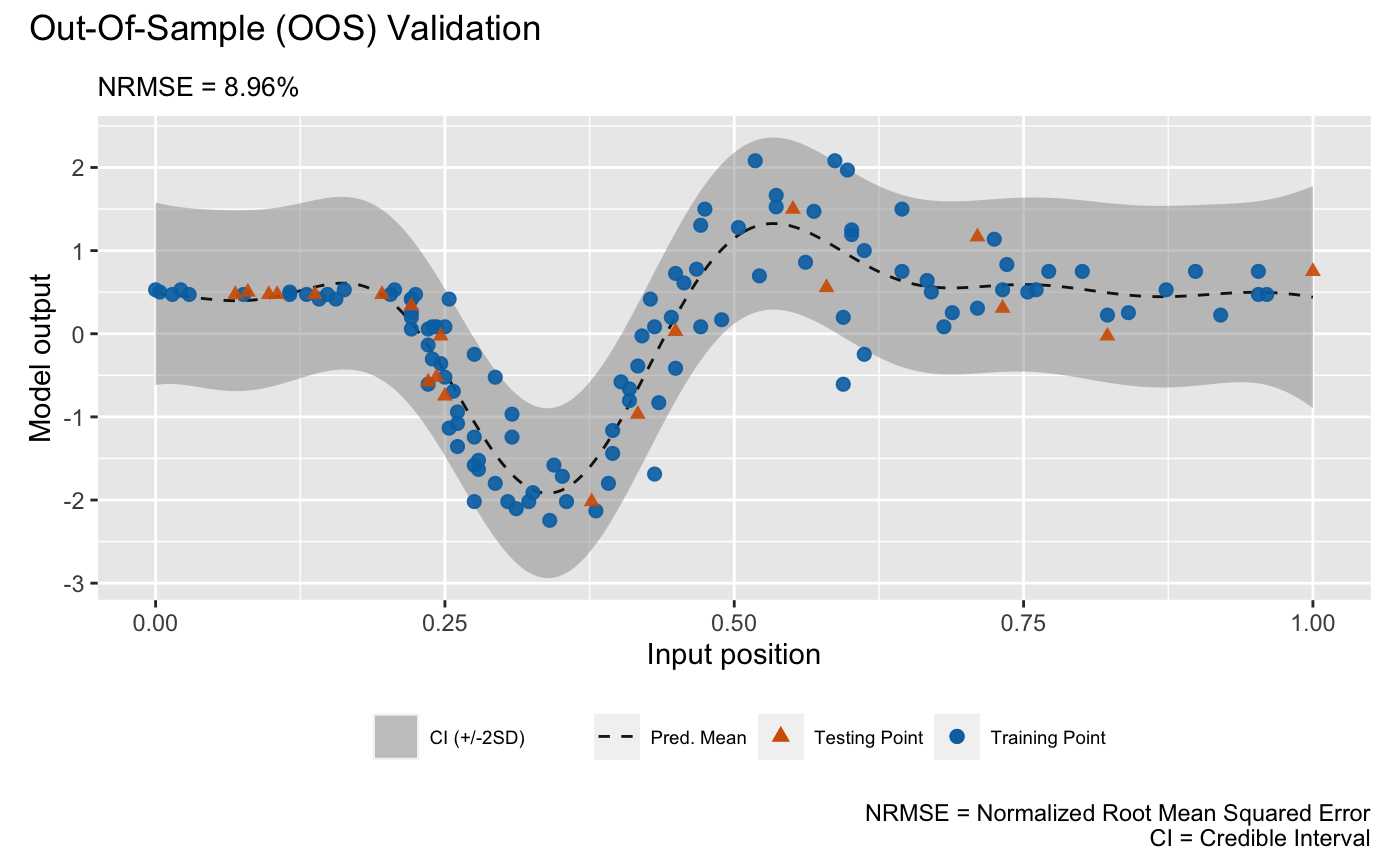
and for the DGP emulator:
plot(m_dgp, test_x, test_y)## Validating and computing ... done
## Post-processing OOS results ... done
## Plotting ... done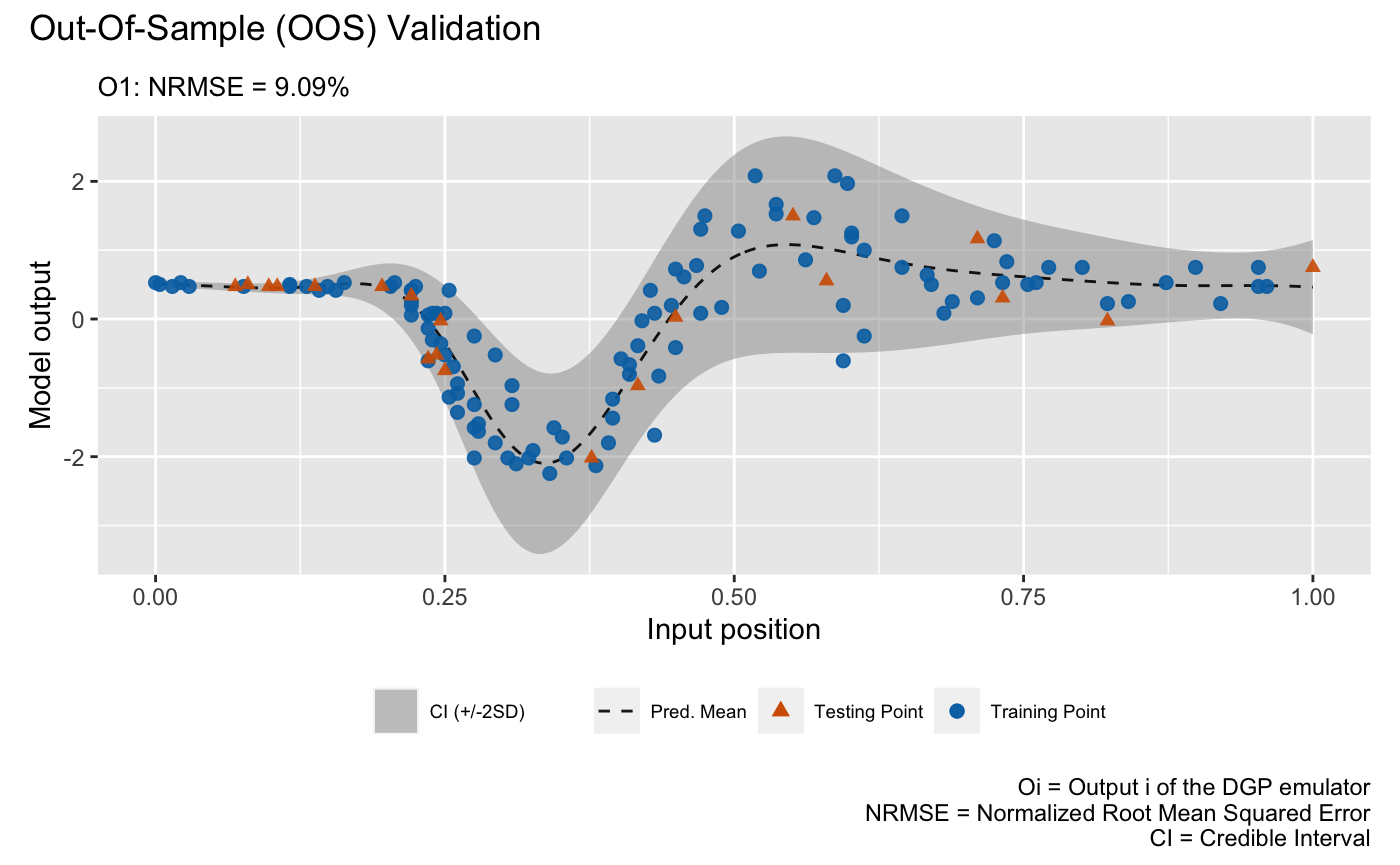
Note that we still need to provide test_x and
test_y to plot() even they have already been
provided to validate(). Otherwise, plot() will
draw a LOO cross validation plot. The visualizations above show that the
DGP emulator gives a better performance than the GP emulator on modeling
the heteroskedastic noise embedded in the underlying data set, even
though they have quite similar NRMSEs.